- Animal Breeding 101
- A Punnett Square A tool to Help Manage Simple Genetic Traits
- Simply-Inherited Trait
- Dominance / Epistasis
- Qualitative vs. Quantitative Traits
Basics of Inheritance
Cattle, like people, inherit half of their genetic makeup from the sire and half from the dam. The genetics are inherited through chromosomes. Cattle have 30 pairs of chromosomes - meaning 60 total chromosomes. Each chromosome is made up of DNA in a long strand that is compacted tightly into the nucleus of each cell. Certain sections of the DNA called “genes” are transcribed into proteins that control nearly every function in life.
Every sperm and egg cell contains a unique set of chromosomes that are combined during fertilization to make a full set with two copies of each chromosome (except for the sex chromosomes) in the newly fertilized egg. Because each sperm and egg cell randomly end up with half of the available genetic material (chromosomes), every sperm and egg cell is unique and every resulting embryo contains a unique combination of genes inherited from its parents. This explains why full siblings can be so different from each other.
A Punnett Square: A tool to Help Manage Simple Genetic Traits
By Jackie Atkins, PhD
A Punnett square is a handy tool to predict expected progeny outcomes from a specific mating. A Punnett square only works with simply inherited traits (where one or just a few genes control the phenotype i.e., coat color, horned/polled, or many genetic conditions). To use a Punnett square, you should know the genes involved with the trait, the genotypes of the parents, and whether certain alleles (variations of the same gene) are dominant, recessive, or somewhere in between.
A few principles that will help you understand how to use a Punnett square:
- All animals have two copies of each gene and pass along one copy to their offspring.
- In a population, there can be several alleles for one gene; but, an individual can have at most two varieties per gene.
- An individual with the same two alleles for a gene is homozygous. An individual with two different alleles of a gene is heterozygous.
- An animal has an equal chance of passing on each one of their alleles.
- In traits with complete dominance (for example coat color), an animal needs only one of the dominant alleles to display the dominant phenotype. An animal needs two recessive alleles to display the recessive phenotype.
- To organize a Punnett square, assign the top of the square for one parent’s genotype and use the left side of the square for the other parent. You will want one row or column per genotype option. The genotype options depend on the potential alleles passed from the parents to the offspring. For example, if a parent has a genotype of Aa, the parent will pass either an A or a to the next generation and these would be the genotype options. If looking at two genes, then all combinations of the alleles for each gene need to be included (an individual that is Aa for gene 1 and Bb for gene 2 could pass AB, Ab, aB, or ab to the next generation). Starting with the top parent, you can fill in each column with the header genotype. Similarly, each row can be filled in with the genotype option listed to the left for that row. Each resulting cell will represent a potential genotype for the offspring (half from the sire and half from the dam) and has an equal chance of occurring.
- If you mate two heterozygous parents for one trait and the trait is only controlled by one gene, you will have a Punnett square with four blocks (two options per parent). If you have a trait controlled by two genes (or if you are looking at the frequency of offspring for two traits) and the parents are heterozygous, then you will have a Punnett square with 16 cells (two different genes and two alleles/gene = four options per parent). See examples below.
- Some of the cells in the Punnett square will have duplicate genotypes and these can be added together to find the predicted frequency of the offspring.
- Some of the cells within the Punnett square will have different genotypes but the same phenotype. For instance, with a completely dominant trait, the heterozygotes will appear the same as the homozygote dominant animals.
Given the above principles, here are examples of using Punnett squares to predict offspring from different matings.
Example 1: Coat Color, a simply inherited trait that exhibits complete dominance.
Alleles: E = black and dominant, e = red and recessive
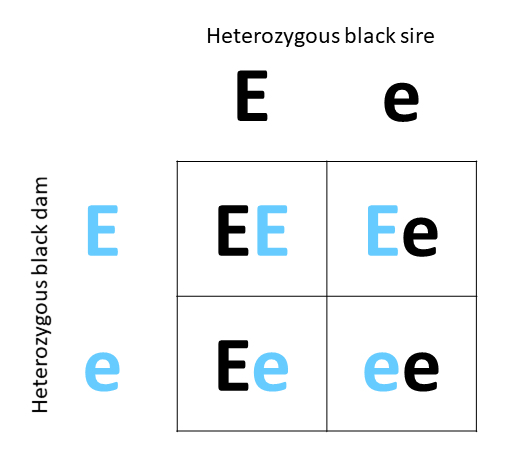
Mating: A heterozygous black sire with a heterozygous black dam
Progeny outcome: In this example, 25% of the calves will be homozygous black (EE), 50% of the calves will be heterozygous (Ee), and 25% of the calves will be homozygous red (ee). As the black allele is dominant and it only takes one black allele to display the black phenotype, 75% of the calves will have a black coat color (EE and Ee calves) and only the homozygous red calves will have red coat color (25%, ee).
Example 2: Two genes that affect the same phenotype: Dilution of Color.
This is an instance where a second gene affects the phenotype of the first gene (a phenomenon called epistasis, see the ASA Science forum post called “Coat Color Dilution in Simmental Cattle” at http://www.simmental.org/forum). The dilution effect is a dominant trait where the dilution allele will cause genotypically black animals to have a grey coat color. Red animals are typically unaffected by the dilutor gene (or possibly slightly lighter in color) but can pass the gene to their offspring.
Alleles: Coat color: E = black and dominant, e = red and recessive
Dilution: D = coat color dilution (if black) and dominant, d = normal coat color and recessive
Mating: The sire is heterozygous for coat color (Ee) and homozygous for the normal dilutor allele (dd). The sire has two potential genotype combinations, Ed and ed. The dam is homozygous red (ee) and carries the dilutor allele (Dd). The dam also has two possible genotype pairings, eD or ed.
P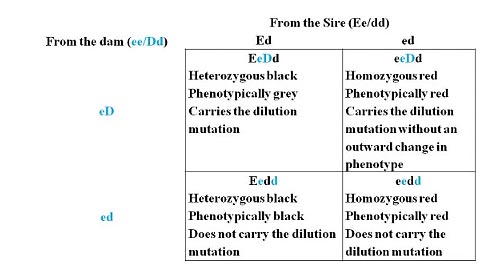 rogeny outcome: Each one of the resulting cells has an equal chance of occurring and each cell is genotypically unique; therefore, each genotype has a 25% chance of occurring (1/4). Phenotypically, we would expect 25% of the calves to be grey (and a carrier of the dilutor allele), 25% black, and 50% red (with half of these carriers of the dilution mutation).
rogeny outcome: Each one of the resulting cells has an equal chance of occurring and each cell is genotypically unique; therefore, each genotype has a 25% chance of occurring (1/4). Phenotypically, we would expect 25% of the calves to be grey (and a carrier of the dilutor allele), 25% black, and 50% red (with half of these carriers of the dilution mutation).
Example 3: Two separate genes with two independent traits, Coat Color and Horned/Polled.
In this example, we will use two heterozygous parents for each trait to display a more complicated Punnett square (16 cells). Neither gene affects the other so the traits are independent of one another. Both traits are completely dominant (black coat color is dominant to red and polled is dominant to horned). In reality, there are other genes that affect these traits (dilution, see above, or the scurred gene in polled cattle, but for this example, we will ignore these other genes).
Alleles: Coat color: E = black and dominant, e = red and recessive
Horned/polled: P = polled and dominant, p = horned and recessive
Mating: The sire and dam are both heterozygous parents for each trait. As these are completely dominant traits, both parents will be black and polled, but carry the red and horned alleles. Each parent will have 4 potential genotype pairings.
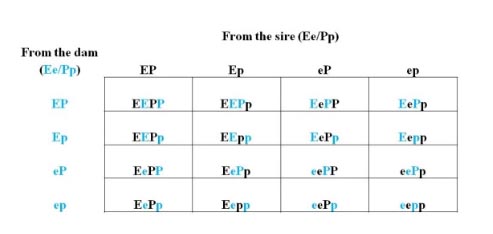 Progeny outcomes: Any calves with at least one E will be black (denoted as E_) and calves with at least one P will be polled (denoted as P_). Out of 16 animals, there will be 9 black and polled offspring (E_/P_; only one homozygous for both traits), 3 black and horned (E_/pp), 3 red and polled (ee/P_), and 1 red and horned (ee/pp) for a ratio of 9:3:3:1.
Progeny outcomes: Any calves with at least one E will be black (denoted as E_) and calves with at least one P will be polled (denoted as P_). Out of 16 animals, there will be 9 black and polled offspring (E_/P_; only one homozygous for both traits), 3 black and horned (E_/pp), 3 red and polled (ee/P_), and 1 red and horned (ee/pp) for a ratio of 9:3:3:1.
These principles can be applied to understanding the frequency of passing along any simply inherited traits including most of the genetic conditions we screen. Combine the information you gain from DNA test results with Punnett squares to help predict potential traits in calves from certain matings. Punnett squares are a useful tool to help make mating decisions to optimize your herd.

Simply-Inherited Trait
Simply-inherited traits are those where one or just a few genes control the phenotype. Coat color, horned/polled status, and many genetic conditions fall under the category of simply-inherited traits. A Punnett square is a handy tool to predict expected progeny outcomes of simply-inherited traits from a specific mating. To use a Punnett square, you should know the genes involved with the trait,
the genotypes of the parents, and whether certain alleles (variations of the same gene) are dominant, recessive, or somewhere in between. Some tips to remember when using a Punnett square:
- All animals have two copies of each gene and pass along one copy to their offspring.
- An individual with the same two alleles for a gene is homozygous. An individual with two different alleles of a gene is heterozygous.
- In traits with complete dominance, like coat color, an animal needs only one of the dominant alleles to display the dominant phenotype. An animal needs two recessive alleles to display the recessive phenotype.
- Some of the cells within the Punnett square will have different genotypes but the same phenotype. For instance, with a completely dominant trait, the heterozygotes will appear the same as the homozygote dominant animals.
Example: Coat color, a simply-inherited trait that exhibits complete dominance.
- Alleles: E = black and dominant, e = red and recessive
- Mating: A heterozygous black sire with a heterozygous black dam
Dominance/Epistasis
Dominance
A dominant trait means an animal needs only one copy of the allele to display the trait. An example of a completely dominant trait would be black coat color. If an animal gets one copy of the black allele and one copy of the red allele, the animal will be black. A recessive trait means an animal needs to inherit two copies of that allele in order to display the trait (phenotype). This is only possible if both parents have at least one copy of this allele.
For example, red coat color is a recessive trait. A calf must have two copies of the red coat allele (ee) to display a red coat. That does not mean both parents had to be red, they just had to carry at least one copy of the red coat allele. Mating two heterozygous black parents (Ee) would have a red calf (ee) approximately 25% of the time.
Dominance is also important in polygenic traits (traits that involve multiple genes like growth). Dominance is the main reason for hybrid vigor or inbreeding depression as increasing genetic diversity leads to an increase in dominant gene expression and a resulting boost in production.
People often equate a dominant trait to mean “good” and recessive to mean “bad”. While this may frequently be the case (for instance, many lethal mutations are recessive), it is not always the rule. As mentioned about the true definition of dominant or recessive traits has nothing to do with good or bad alleles, just how many copies are needed to display the phenotype.
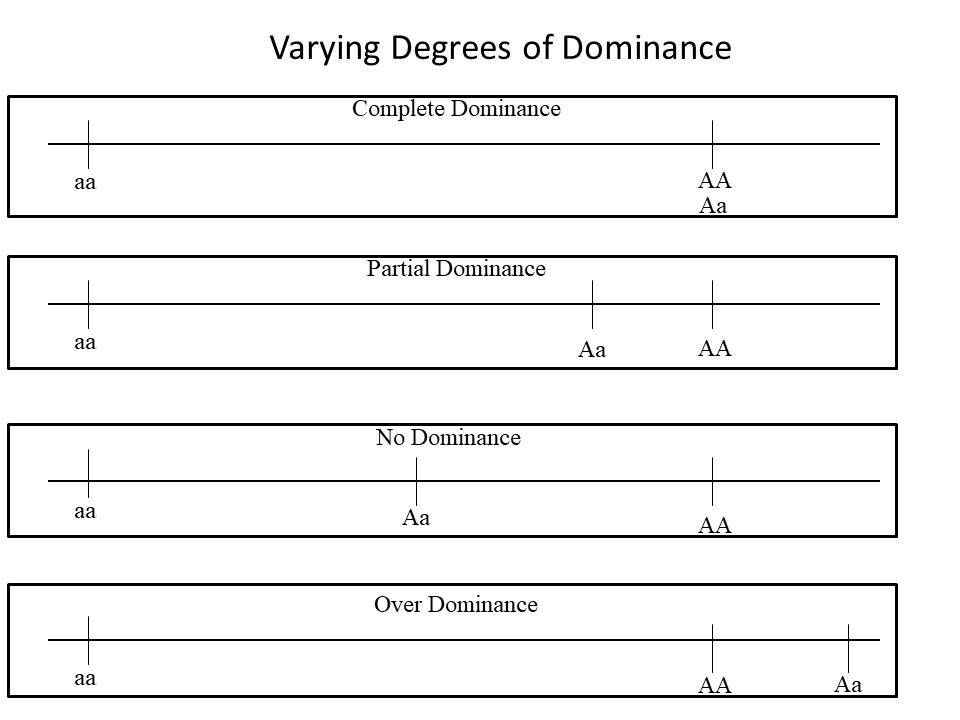
Varying Degrees of Dominance
Frequently when we talk of dominant traits, we automatically think of completely dominant traits where an animal with two copies of the dominant allele is phenotypically the same as an animal with just one copy of the dominant allele. This is the case with black coat color or the polled trait in cattle. However, many traits have varying degrees of dominance where the recessive allele is not completely masked by the presence of the dominant allele. Here we provide definitions of the varying degrees of dominance (complete, partial, no dominance, overdominance) and a figure to illustrate each form.
Complete dominance: The dominant allele completely masks the recessive alleles resulting in a heterozygote that is phenotypically the same as the dominant homozygotes.
Partial dominance: One allele is not completely dominant over the other allele. With partial dominance, the heterozygote has a phenotype between the two homozygotes but more similar to the homozygote dominant animals.
No dominance: The heterozygote exhibits a phenotype that is exactly in between the homozygotes.
Overdominance: The heterozygote exhibits a phenotype that is beyond the range of either of the homozygotes but is more similar to the dominant homozygotes.
Caption: Illustration of varying degrees of dominance. The horizontal lines represent a phenotypic outcome of the genotype for a particular gene. “A” represents the dominant allele and “a” represents the recessive allele. Note that in all four examples the homozygotes have the same phenotypic outcome, what differs between various types of dominance is the phenotype of the heterozygotes.
Example: Consider a theoretical gene that influences growth up to weaning. Assume the dominant dominant homozygous animals have a 5 lb advantage at weaning and the homozygous recessive animals have 0 lb advantage from this one gene. With varying degrees of dominance, you could expect the following for the heterozygous animals:
Complete Dominance - 5 lb advantage (same as homozygous dominant)
Partial Dominance - 3 lb advantage (not the same as homozygous dominant but closer to it than average)
No Dominance - 2.5 lb advantage (average of the dominant and recessive homozygotes)
Over Dominance - 7 lb advantage (beyond the homozygote dominant animals)
Epistasis
Epistasis refers to the interaction among genes at different locations of the chromosome. The resulting phenotype from the gene at one location is determined by the gene at another location. The polled gene and phenotype is an example of epistasis where a second gene (scurred) affects the phenotype of the polled animal. The presence of scurs (typically small, movable, hollow pseudo horns) only occurs in heterozygous polled animals. All homozygous polled (PP) animals are phenotypically polled independent of the scurred gene. All homozygous horned (pp) animals have horns and are unaffected by the scurred gene. The heterozygous animals at the horned/polled gene (Pp) can either be polled or have scurs depending on their sex and the alleles for the scurred gene. Similar to the varying degrees of dominance, epistasis also contributes to inbreeding suppression and hybrid vigor.
Qualitative vs. Quantitative Traits
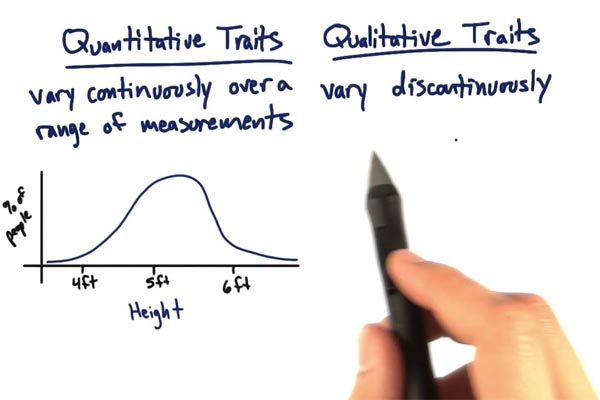
A qualitative trait is a trait that can be described as a category. For example, black or red coat color, horned or polled, coat color dilution are all qualitative traits. Qualitative traits are frequently controlled by one or just a few genes meaning they are simply-inherited traits. Conversely, quantitive traits are traits with phenotypes that can be measured numerically. These are traits like weaning weight and dry matter intake. Usually, quantitative traits are polygenic meaning they are controlled by many genes and gene interactions. Some traits called threshold traits, are polygenic but expressed in categories - pregnant/open and occurrence of dystocia for example.
Qualitative traits often (not always) have DNA tests for the genes of influence and breeders can use a Punnett square to determine the likelihood of different calves from a certain mating. Quantitive traits typically require EPDs to make progress in genetic selection in those traits.
Animal Breeding
History of Cattle Breeding
The history of breeding cattle in the United States is very dynamic. Breeders have gone through many stages and changes, each built from previous techniques and knowledge. Although past stages focused heavily on popular folklore, today’s focus is built on a foundation of sound science for producer profitability and sustainability.
Video on the History of Cattle Breeding by beefreproduction.org.